Nota Bene: The softclipping and full CIGAR string generation described in this document is currently under a PR for inclusion into a point release of salmon in the near future. For the time being, to use / test this feature, you will have to build salmon from the softclip branch. This document will be updated again when the feature has been integrated into a tagged release of salmon.
In this tutorial, we show how to use the improved soft-clipping feature of Salmon and some new features such as CIGAR computation for SAM/BAM alignment outputs.
Why does soft-clipping matter?
Soft-clipping in alignments can be very useful. It allows the alignment to be cut at the beginning or the end of a read without a penalty. Therefore, it can be comprehended as local alignment of a read to a target sequence instead of an end-to-end alignment.
One practical use case of soft-clipping / local alignment is to give the aligner module the power to trim the adapter from either side of the sequence reads.
Another use case is when the read sequence is overhanging the target sequence on either 5’- or 3’-end. It is also possible that the read sequence overhangs from both ends of the target sequence if the target sequence is shorter than the read sequence. In this case, if we align the read in an end-to-end fashion, the alignment will have insertions in the prefix and/or suffix of the read in the overhanged sections. This could cause the read alignment to have a high penalty and a very low score. In particular, in Salmon such alignments might end up getting rejected if their score drops below the minimum accepted score, minScoreFraction * maximum_achievable_score where maximum_achievable_score = read_len * match_score. Thus, in such cases, we would like to avoid penalizing the overhanging prefix and/or suffix of the read to get a more realistic alignment score.
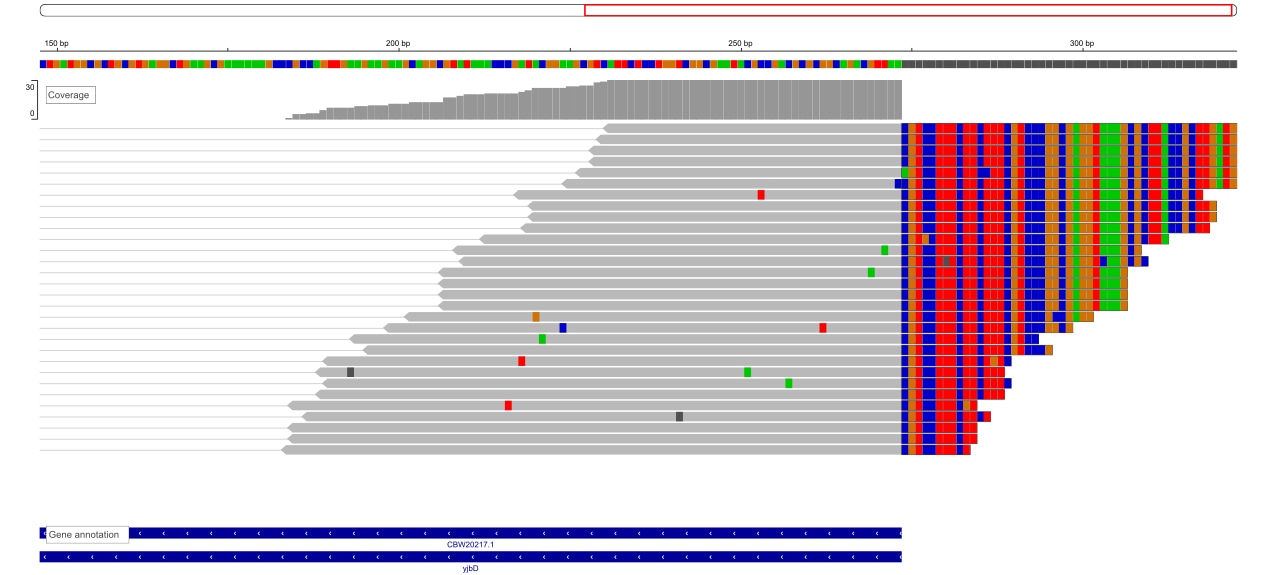
The figure below shows an example of overhanging soft-clipped alignments in IGV genome browser. Notice that the coverage track on top of the alignments track is cut at the 273rd base which is exactly the end position of the gene. For better demonstration and in order to show the full-length alignments, the “show soft clips” flag is enabled in the genome browser. Thus, the soft-clipped portion of the reads are colored in every position based on their nucleic acid type. Note that the soft-clipped portion is not counted towards the coverage of the region.
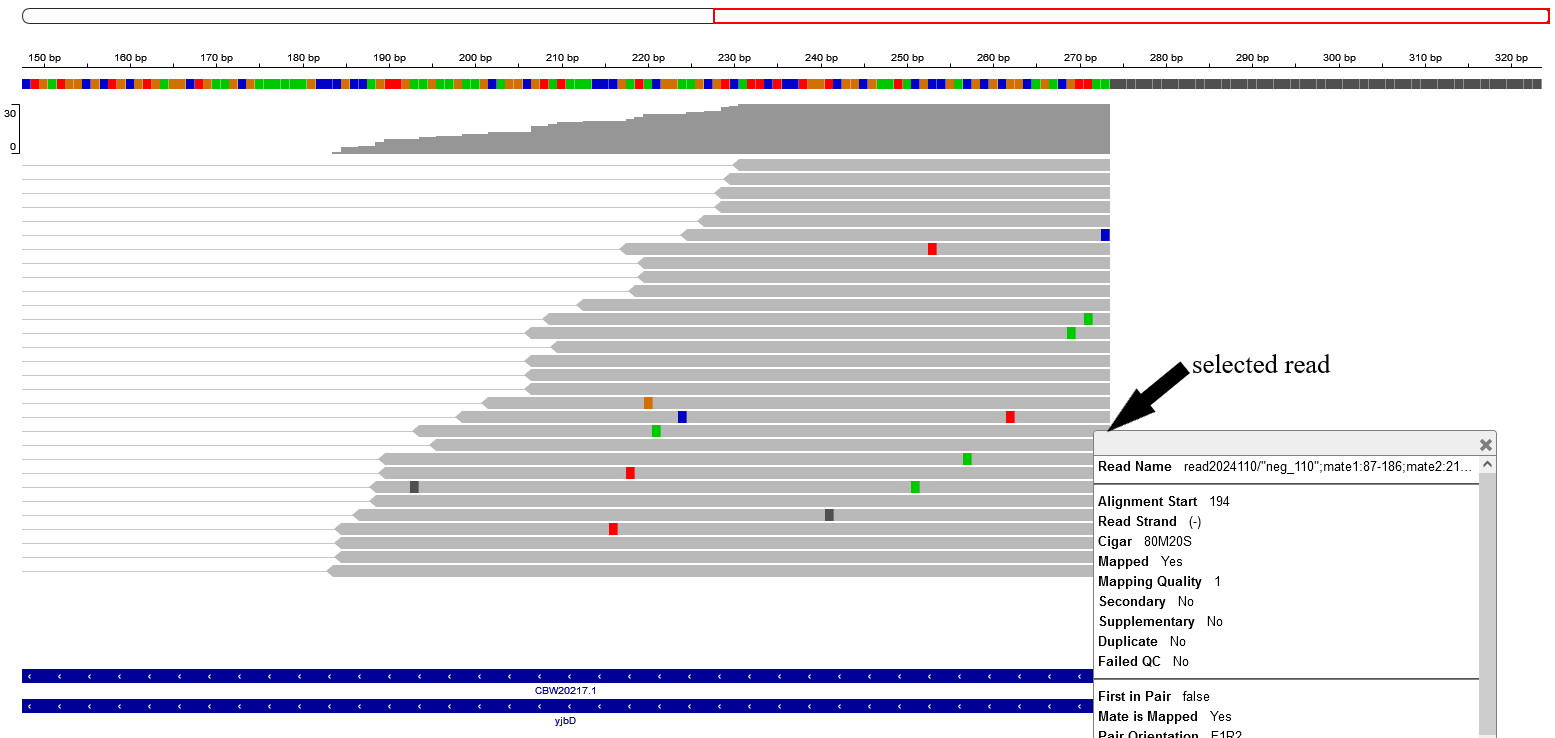
In the next figure, the soft-clipped bases are hidden and as an instance. As an example, for the alignment selected below the CIGAR string (80M20S) means that the last 20 base-pairs of this particular read are soft-clipped.
Why does soft-clipping affect Salmon quantifications? And how does it improve it?
Enabling the soft-clip feature in Salmon can directly affect the results of its gene / transcript quantification method. The reason is that the score of a soft-clipped read alignment is most likely different from its end-to-end alignment. That’s because the soft-clipped portion of the read is not penalized unlike an end-to-end alignment where every base should either be a match, mismatch, insertion or deletion. The change in the alignment score could affect the decision on whether to count the alignment towards its mapped gene / transcript as well as the probability of the confidence of this mapping.
In some cases, the soft-clipped alignments are more accurate, especially in the cases mentioned above, including the over-hanged alignments and adapter trimming. Thus, the quantification results could be improved by enabling soft-clipping in Salmon.
How to enable soft-clipping in Salmon?
By default, Salmon selective-alignment performs an end-to-end alignment. In order to enable soft-clipping in Salmon, one should pass --softclip flag in the quant command of Salmon. Since Salmon only performs the read mapping procedure in the selective-alignment mode, the --sofclip flag is only compatible in that setting.
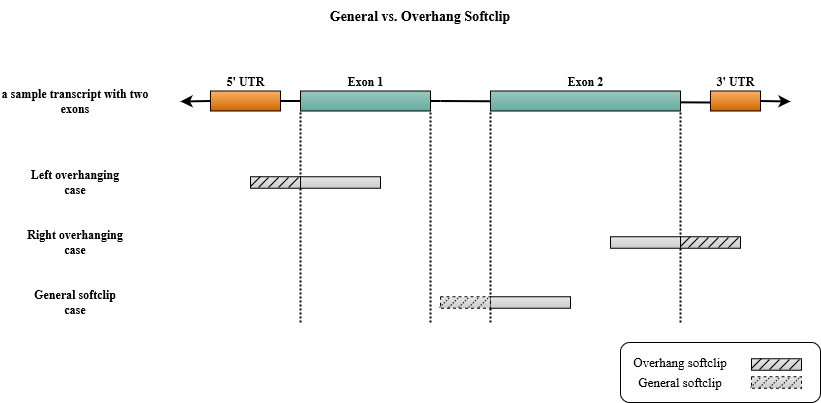
The user is able to control the rate of the soft-clipping using --maxSoftclipFraction parameter which restricts the fraction of the soft-clipped part of a read. An alignment is accepted and reported if and only if alignment_score >= minScoreFraction * max_alignment_score where max_alignment_score = match_score * (read_length – softclip_length). This threshold applies separately to the mates of a paired-end read and the soft-clip length on a mate will not affect the maximum allowed soft-clip length on the other mate. In salmon, this parameter can accept either one or two values. If two values are passed, they will specify the maximum soft-clip fraction for the general cases and transcript overhanging cases respectively. If one value is passed, the same threshold will be used for general and overhanging soft-clips. This means that you are free to choose different thresholds for overhanging soft-clips and general soft-clips that happen in the middle of a target reference. This way, even if you allow large overhanging soft-clips, you can restrict the general soft-clips by setting a smaller value as their maxSoftclipFraction to avoid spurious mappings in the middle of the target sequences. It can be especially useful for quantification of bacterial transcriptome where the target sequences are typically short and large overhanging soft-clips are frequent. The figure below illustrates an example of general and overhanging soft-clipped alignments on a sample transcript with two exons. The overhanging cases are happening in the beginning or the end of the transcript when the rest of the alignment is getting out of the reference transcript, while the general soft-clip cases could happen in the middle the reference. In this particular example, the reason behind the general soft-clip case could be intron retention.
Note that if the softclip flag is passed to the program, the default value for maxSoftclipFraction would be 0.2. However, you can set it to any value from 0 to 0.8. This means that at most 80% of a read can get soft-clipped in Salmon.
If the total length of the soft-clipped regions on a read (the summation of the soft-clip lengths in the beginning and the end of the read) is greater than maxSoftclipFraction * read_length, soft-clipping will not be permitted; however, another chance will be given to the read to be aligned end-to-end. If the end-to-end alignment is acceptable based on its score, it will be reported and considered in the quantification step. For more information regarding the Salmon parameters, please refer to the Salmon documentation. For the selective-alignment mode of quant, you can get the help message using the following command:
salmon quant --help-reads
By passing –softclip flag, soft-clipping will be enabled:
salmon quant -i salmon_index -la -r reads.fq -o sc_res -p 8 --softclip
Here, --maxSoftclipFraction is not passed so the maximum allowed soft-clip ratio is set to 20% by default, which means no soft-clipping greater than 20% of the read length will be accepted.
- Note 1. The
--softclipOverhangsflag is deprecated and it is set to have the exact same effect as the--softclipflag. However, you can control the soft-clip rate separately for general and overhang cases using the--maxSoftclipFraction. For example, when soft-clipping is enabled, to prohibit general soft-cliping while allowing at most 30% of the read to be overhanging, you should pass--maxSoftclipFraction0 0.3. - Note 2. In the mapping mode of Salmon, the match score, mismatch penalty, gap open and gap extend values can be changed by the user in special situations; however, you most likely don’t need to specify the values for these parameters since the default values are carefully selected to work best in many cases. Another mapping parameter is the
--endBonusparameter which is newly added to Salmon and specifies how strict the soft-clipping decisions are made in the program. It specifies the extra bonus score that is added to the end-to-end alignment score, when the alignment reaches the end of query. The larger this value is set, the more likely that soft-clipping is not reported as part of the alignment of a particular query because getting to the end of the query would be preferred instead.
How to enable accurate CIGAR computation in Salmon?
The alignment results of the selective-alignment mode of Salmon can be reported in SAM format. However, by default, the CIGAR string of the SAM record is approximately calculated to reduce the total running time. You will be able to change this behavior in case you need the accurate CIGAR strings to be reported in the SAM records. To do so, you need to pass --computeCIGAR flag to the quant command of Salmon in the selective-alignment mode.
For example, the following end-to-end selective-alignment command will generate accurate CIGAR strings in the SAM output:
salmon quant -i salmon_index -la -r reads.fq -o e2e_res -p 8 --computeCIGAR --writeMapping mappings.sam
Example 1 – Adapter trimming
In this example, we look at a dataset that in some of its reads, the adapters are attached to the biological sequences in the input file. A common practice is to use a sequencing trimming method to remove the adapters from the reads so that the quantification is then performed on the biological sequences only. Here, we perform the quantification in both trimmed and original reads. The sample that we work on in this example is a FFPE RNA-Seq sample which is publicly available on SRA under SRR5314081 tag.
In order to perform quantification in selective-alignment mode in Salmon, we need to build an index on human transcriptome. Here we use ensemble v101 human transcriptome. To download the transcriptome and build salmon index:
wget http://ftp.ensembl.org/pub/release-101/fasta/homo_sapiens/cdna/Homo_sapiens.GRCh38.cdna.all.fa.gz
salmon index -t Homo_sapiens.GRCh38.cdna.all.fa.gz -i salmon_index -p 8
This will create a directory named “salmon_index” containing the Salmon index files using 8 threads. Then, we perform the quantification on the trimmed dataset.
salmon quant -i human_index -la -1 reads_trim_R1.fq -2 reads_trim_R2.fq -o trimmed_e2e_res -p 8 --seqBias --posBias --thinningFactor 64
Now, we run Salmon quantification without enabling the soft-clipping on the original dataset that has the adapters attached to the read sequences. This means that Salmon will perform end-to-end alignments.
salmon quant -i human_index -la -1 reads_R1.fq -2 reads_R2.fq -o orig_e2e_res -p 8 --seqBias --posBias --thinningFactor 64
Finally, we run the original dataset with adapters included again but this time we enable the soft-clipping.
salmon quant -i human_index -la -1 reads_R1.fq -2 reads_R2.fq -o orig_sc_res -p 8 --seqBias --posBias --thinningFactor 64 --softclip
We can now compare the correlation of the three resulting quantification matrices which you can find in the table below using Spearman correlation methods. As you can see, the correlation of the trimmed quantification matrix is closer to when we enable soft-clipping on the original dataset rather than performing an end-to-end global alignment. The reason is that soft-clipping acts as a trimming method in this case and ignores the adapter sequences in the beginning or end of the reads in case of many mismatches. Thus, the achieved alignment scores will be more realistic and closer to when the adapter sequences are completely removed (as in the first run).
| Trimmed end-to-end | Original end-to-end | Original soft-clip | |
|---|---|---|---|
| Trimmed end-to-end | 1.0000 | 0.9212 | 0.9458 |
| Original end-to-end | 0.9212 | 1.0000 | 0.8945 |
| Original soft-clip | 0.9458 | 0.8945 | 1.0000 |
Example 2 – Prokaryotic expression quantification
In this example, we look at another use-case in quantification of bacterial transcriptome. Most bacteria lack annotated transcriptomes and generally, the quantification is performed on annotated CDS regions instead. These genes are typically grouped into co-transcribed regions called operons as shown below.
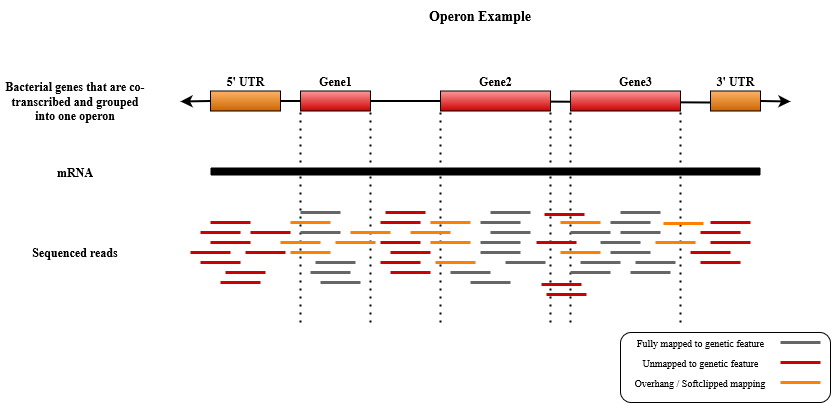
Here, we use a dataset of paired-end RNA-Seq short reads simulated from Salmonella. We process the dataset using two different representations of Salmonella transcriptome. The first one is the regular gene/CDS-based transcriptome that is commonly used in prokaryotic expression quantification. The second simulated transcriptome is an operon-based transcriptome in which the sequences of the co-transcribed genes (e.g., gene 1, 2, and 3 in the figure above) are concatenated together to form a united operon sequence. The simulated data and transcriptome reference files can be downloaded here.
In order to show the effectiveness of soft-clipping reads that overhang a gene / CDS we build two different indexes. One using the operon-based transcriptome as a ground truth and another index using the commonly used gene-based transcriptome.
To build the indexes:
salmon index -t operons.fasta -i operon_index -p 8
salmon index -t genes.fasta -i gene_index -p 8
Now we can run the quantification command on a paired-end simulated dataset using these two indexes and different settings for soft-clipping to show the effect of it.
For operon index we run quantification in end-to-end mode as well as soft-clipping mode with different maximum allowed soft-clip threshold (20% and 60%):
salmon quant -i operon_index -la -1 reads_R1.fq -2 reads_R2.fq -p 8 --seqBias --posBias --thinningFactor 64 -o ob_e2e_res
salmon quant -i operon_index -la -1 reads_R1.fq -2 reads_R2.fq -p 8 --seqBias --posBias --thinningFactor 64 --softclip --maxSoftclipFraction 0.2 0.2 -o ob_sc_0.2_res
salmon quant -i operon_index -la -1 reads_R1.fq -2 reads_R2.fq -p 8 --seqBias --posBias --thinningFactor 64 --softclip --maxSoftclipFraction 0.6 0.6 -o ob_sc_0.6_res
We run the same commands but this time in gene-based mode:
salmon quant -i gene_index -la -1 reads_R1.fq -2 reads_R2.fq -p 8 --seqBias --posBias --thinningFactor 64 -o gb_e2e_res
salmon quant -i gene_index -la -1 reads_R1.fq -2 reads_R2.fq -p 8 --seqBias --posBias --thinningFactor 64 --softclip --maxSoftclipFraction 0.2 0.2 -o gb_sc_0.2_res
salmon quant -i gene_index -la -1 reads_R1.fq -2 reads_R2.fq -p 8 --seqBias --posBias --thinningFactor 64 --softclip --maxSoftclipFraction 0.6 0.6 -o gb_sc_0.6_res
Note that the summation of TPM for the genes that are grouped in an operon is calculated so that the matrices become comparable between gene-based and operon-based quantification results. The python script below performs this conversion:
# convert.py:
import sys
import re
import argparse
parser = argparse.ArgumentParser(description="Converts gene based quantification to operon based quantification")
parser.add_argument("--gff", required=True, help="annotation of operons and genes")
parser.add_argument("--quant", required=True, help="salmon quantification file")
parser.add_argument("--output", required=True, help="output salmon quantification")
params = parser.parse_args()
# load the gene to operin assignments from the input gff file into gene2operon dictionary
gene2operon = {}
with open(params.gff) as fin:
for line in fin:
l = line.strip().split()
anno = re.split(";|=", l[8])
genes = []
operon = ""
for i in range(len(anno)):
if anno[i] == "transcript_id":
operon = anno[i+1]
if anno[i] == "operon_gene":
genes.append(anno[i+1])
for g in genes:
gene2operon[g] = operon
# load the input gene-based quantification result and do the conversion to operon-based
header = ""
operon_quant = {}
gcnt = 0
with open(params.quant) as fin:
first = True
for line in fin:
if first:
first = False
header = line
continue
gcnt += 1
l = line.strip().split()
gene = l[0]
vals = [float(v) for v in l[1:]]
operon = gene2operon[gene]
if operon in operon_quant.keys():
(operon_q, cnt) = operon_quant[operon]
operon_q = [a + b for a, b in zip(vals, operon_q)]
cnt += 1
operon_quant[operon] = (operon_q, cnt)
else:
operon_quant[operon] = (vals, 1)
print('{} genes to {} operons'.format(gcnt, len(operon_quant)))
# write the calculated operon-based results in the output file
with open(params.output, "w") as fout:
fout.write(header)
for op in sorted(operon_quant.keys(), key = lambda x: (x[1:-1].split('_')[0], -1*int(x[1:-1].split('_')[1])), reverse=True):
(opq, cnt) = operon_quant[op]
tlen = opq[0]
etlen = opq[1]
tpm = opq[2]
numr = opq[3]
fout.write('{}\t{}\t{}\t{}\t{}\n'.format(op, tlen/cnt, etlen/cnt, tpm, numr))
To run this script, we pass the gene annotation file salmonella_operon.gff which can be downloaded from here.
wget https://zenodo.org/record/5778127/files/salmonella_operons.gff
python3 convert.py --gff salmonella_operon.gff --quant gb_e2e_res/quant.sf --output gb_e2e_res/quant.operon.sf
python3 convert.py --gff salmonella_operon.gff --quant gb_sc_0.2_res/quant.sf --output gb_sc_0.2_res/quant.operon.sf
python3 convert.py --gff salmonella_operon.gff --quant gb_sc_0.6_res/quant.sf --output gb_sc_0.6_res/quant.operon.sf
Now, we can look at the Pearson correlation matrix that is formed from pairwise correlation of the 6 expression matrices (TPM) we collected by running the commands above. Since the simlulation is designed in a way that most of the reads are fully aligned to the operons, the soft-clip feature is not affecting the quantification results. Thus the correlation between different operon-based runs is very close to 1 and omitted from the table. The correlation of the end-to-end operon-based results with the gene-based results is shown in the table below:
| Gene based e2e | Gene based sc_0.2 | Gene based sc_0.6 | |
|---|---|---|---|
| Operon based e2e | 0.3858 | 0.5696 | 0.6859 |
where e2e is short for end-to-end, and sc for soft-clip with different maxSoftclipFraction thresholds. As demonstrated in the table above, allowing soft-clip alignments for gene-based transcriptome increases the correlation to operon-based transcriptome which is used as a ground truth here. In addition, increasing the maxSoftclipFraction increases the correlation even more because in bacterial genomes, the genes and CDS regions can be short which causes overhanging of the reads off the sides of the gene / CDS. Another observation is that enabling soft-clipping for the operons that have the entire co-transcribed sequence do not change the expression quantification matrix and has the highest correlation. Thus, enabling the soft-clipping will not reduce the quality of the quantification.
Running time performance notes
We have benchmarked different datasets for the purpose of examining the effect of soft-clipping and accurate CIGAR computation in the running time of Salmon which proves to be a small effect. On average, enabling the soft-clipping feature in Salmon has 3% overhead in wall clock time. Besides, asking for accurate CIGAR computation in Salmon has 1-2% overhead in wall clock time and if both features are enabled, the running time will have at most 5% overhead.
